Juexin Wang
Building upon previous work on transcriptomics analysis, Luddy Indianapolis BioHealth Informatics assistant professor Juexin Wang and co-investigators have developed a novel AI-based methodology for improving gene expression analyzation in situ. Their findings were published in the November 14 issue of Nature Communications and subsequently chosen as a featured editorial by the esteemed journal.
Artificial Intelligence (AI) has reshaped science with mathematical modelling in biological research and data analytics. Driven by big data from sequencing, AI technologies penetrate the interdisciplinary research in bioinformatics studies, which helps to disclose the hidden patterns of diseases, discover the underlying biological mechanisms, and propose new hypothesis of pathogenesis.
The team’s novel big-small patch (BSP) non-parametric computational model reliably compares expression patterns and identifies spatially variable genes (SVG) in both 2D and 3D. This is an improvement on prior 2D pattern recognition and is previously unreported for 3D pattern recognition. Data from identified SVGs can then be used to link molecular cell function with tissue phenotypes in disease studies.
The BSP algorithm has been shown to successfully and reproducibly identify spatially variable transcriptomes in kidney cells in situ, or ‘living in place’, which allows for further exploration of specific targets for treatment.
Not all cells produce the same transcripts or messenger RNA—even within the same tissue or organ—due to molecular-level errors and modifications, leading to variability in expression. Traditional in vitro methods could not differentiate between healthy and diseased cells harvested from the same tissue sample since the integrity of the layers was compromised or destroyed on removal. Being able to differentiate between normal and abnormal cells and link function to phenotype offers the potential of developing very specific, targeted treatments for disease.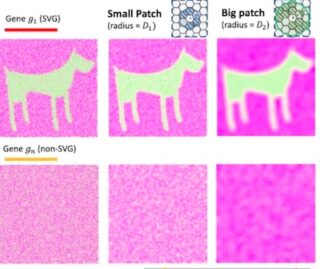
Michael Eadon, a physician at IU School of Medicine and co-author on the Nature publication, provided kidney data and pathological analysis needed to validate the team’s method. “His indispensable biological support validates our computational framework, and makes this interdisciplinary work happen,” Wang said.
Josette Jones, chair of the Department of BioHealth Informatics, said, “Juexin Wang’s research on single-cell sequencing technologies explores the heterogeneity and importance of each cell’s multi-omics. The research not only refines technological approaches to single-cell sequencing but opens up improved diagnostic and treatment options for oncology, neurology, reproduction, immunology, digestive, and urinary tract diseases.”
Acknowledgement
Wang’s research is supported by National Institutes of Health grant R01DK138504 and the AnalytiXIN initiative.
Media Contact
Joanne Lovrinic
jebehele@iu.edu
317-278-9208